Site navigation:
PyDSTool is a Python-based Dynamical Systems Toolkit. With PyDSTool we aim to provide a powerful suite of computational tools for the development, simulation, and analysis of dynamical systems that are used for the modeling of physical processes in many scientific disciplines, but especially in the biological sciences. We place emphasis on the support of data analysis and model fitting as a core part of the process of data-driven modeling. Our focus is on models involving ordinary differential equations (ODEs), differential-algebraic equations (DAEs), and discrete maps. Our approach differs from many existing software packages because of our focus on making model building, simulation, and analysis intuitive and efficient for an advanced end-user (who is unlikely to be either a mathematician or a computer programmer by trade) without sacrificing speed. We say for advanced users because PyDSTool does not provide a graphical interface, but instead a scripting environment with flexible, programmable library tools.
The software package is written primarily in the high-level interpreted language Python, and is freely available under the BSD open-source license from the SourceForge community website and repository. For certain computationally intensive tasks, code written in C and Fortran can be invoked (without interference or contribution at this level by the user).
In pursuing our aims we have developed a hierarchy of object-oriented programming classes available to the modeler. These classes provide a means for “data abstraction”, that is, to provide the user with intuitive “objects” that encapsulate common data structures and methods for manipulating that data through a well-defined interface. These classes naturally make the software highly modular, and they range from defining hybrid model simulators, to individual data points, to symbolic mathematical expressions, to bifurcation analyzers. Some of these classes are built over classes exported by SciPy. We believe that many of these classes have wider scientific utility.
Disclaimer: The PyDSTool software is "research code" in a Beta stage of development, and should not be treated as a complete or comprehensive dynamical systems package, with the associated expectation that its design and implementation have thoroughly stabilized and have been well tested. We have added features as and when we have had a use for them in our own research, and have omitted many important features that we would love to add if time permits us, or if our research so demands.
You might like to submit feature requests, or you may also like to contribute to the code yourself. We are also interested to hear your opinions about the possibility of adding some of our classes to SciPy (perhaps in modified form). Please contact us at the SourceForge open discussion forum or via email.
A dynamical system is defined by a set of rules or transformations for determining how points in a multidimensional space move in time. Time may be either discrete or continuous. The traces of the points as they move in time are called trajectories. The goal of Dynamical Systems Theory is to provide a comprehensive description of the geometric structures arising from these trajectories. In addition to elucidating the dynamics associated with an individual dynamical ssystem, bifurcation theory may be used to describe how the dynamics of a system varies with changes in parameter values.
Interactive numerical and graphical exploration are important tools in dynamical systems research for several reasons:
Consequently, there is a critical need for computational environments that provide effective tools for exploring dynamical systems with minimial effort on the part of the user. The PyDSTool project attempts to provide such an environment for use by both casual and advanced practitioners of Dynamical Systems Theory. We consider only finite dimensional systems (Ordinary Differential Equations, Differential-Algebraic Equations, mappings) and some forms of infinite dimensional delay systems (also see pydelay). We do not consider spatially-extended infinite dimensional systems such as Partial Differential Equations, for which other packages exist. We also do not focus on the kinds of heterogeneous "hybrid" systems that are primarily defined by discrete event simulation, as used extensively in process control and engineering applications (for instance see SimPy).
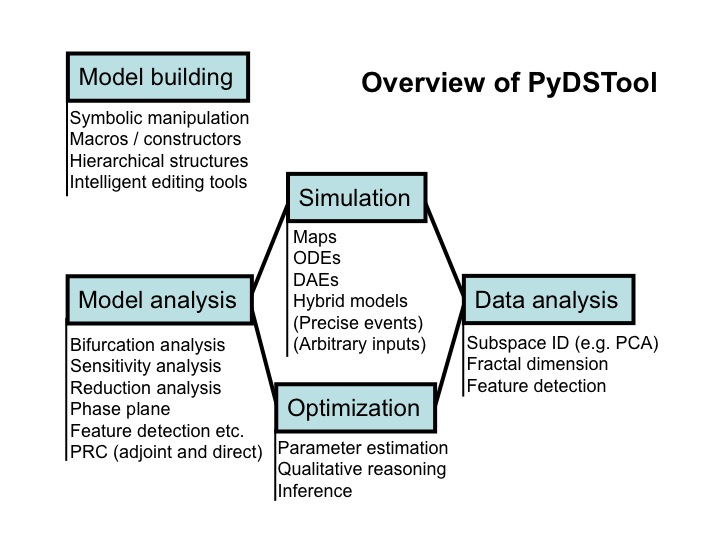
A summary of the main features and application areas of the toolboxes is given in the diagram below.

Our package incorporates:
Efficient and state-of-the-art ODE / DAE / discrete map simulation tools (using dynamically-linked and automatically generated C code, if external compiler available) (see Generators)
Hybrid model and event-driven simulation support (see HybridSystems)
Simulations and analysis can be forced to be "bounds safe", e.g. for "non-negativity preservation" (see BoundsSafety)
Bifurcation analysis and continuation tools in-built, via PyCont with AUTO
Interactive command-line / script-based interface
"Index-free" and context-heavy data structures, including an enhanced version of arrays (see Pointsets)
Symbolic expression utilities (including evaluation, substitution, derivatives, some simplification) (see Symbolic)
Easy to build complex models using hierarchical object-oriented data structures that contain composable model specifications (see ModelSpec)
Memory management utilities, data import and export (including SBML conversion and LaTeX markup via the SloppyCell package, see Section 10 of the user documentation)
Modular code design allows easy expansion to support other algorithms (contributions welcome)
Data structures and toolkits for parameter estimation / model fitting ("constraint based modeling") and other time-series and data-driven problems (see Parameter Estimation)
Seamless use with tools in SciPy, NumPy, etc. through dynamic typing
Additional toolboxes for specific applications, including biomechanical modeling, computational neuroscience, and systems biology (see ToolboxDocumentation)
Most of these features are simply not available in the native SciPy ODE integrators or in application-specific solvers.
Users of PyDSTool need to be familiar with working in interactive, command-line environments such as UNIX or Matlab, including the writing of command scripts. There is presently no graphical interface for PyDSTool. Our emphasis is on the interactivity of a command-line and the rapid prototyping possibilities of script-based computing and a programmatic interface. Although there is an initial learning curve, the opportunities for sophisticated calculations are much greater with a programmatic interface than a graphical one.
In building a core library of Python classes, supporting many fundamental concepts in dynamical systems modeling, we provide more than just a glue with which to interface multiple tools. Our basic object classes store rich "context" that carries a lot of useful mathematical baggage. Through interaction with our Python environment at the script level, users can build complex models in a structured way, and have access to mathematically intuitive information about the models, using the intrinsic context of all the Python objects at the heart of their computations. Thus, our user interface model is based on users interactively "querying" (a.k.a. "introspecting") objects for their attributes and properties. The layers of class abstractions in PyDSTool frees users from dealing with the usual low-level coding details of simulation and data manipulation, and empowers them with mathematically intuitive tools to express their specifications and design their computations in high level, intuitive terms. This also eases the overhead in interfacing the basic numeric classes and simulators with tools and utilities such as parameter estimators or data analyzers.
We believe it is crucial for users to be able to combine the application of tools in a nested or interleaved fashion, in order to make the most flexible and dynamic manipulations of a model. Such rich combinations may be difficult or imporssible in disjointed software environments, and we believe our community is eager to be able to smoothly set-up and maintain such situations for their own modeling projects. It is a challenge to cleanly and efficiently interface different legacy algorithms with the core Python code in order to maximize the use and re-use of the context associated with the core objects.
Users are provided with an interface for the specification of both simple and complex dynamical systems models, using minimal programming syntax, and a range of options in converting these abstract specifications into instantiated numerical solvers for a specific system. Within the same interactive session, users have immediate access to analysis tools for continuation, parameter estimation, optimization, and so on. These tools are each tailored for use with the core PyDSTool structures to ensure the user has to write as little additional computer code as possible. Extensive documentation for the project has been provided online on this wiki.
A key aspect in the design of PyDSTool is the provision of adequate diagnostic information and querying utilities for data structures and computations. Users can expect helpful information regarding the status of their model development and computations beyond the guidance of the online documentation, through in-built querying commands and detailed error messages. The object-oriented nature of the software also provides inherent protection of the users’ conceptualization of data-flow and control in their PyDSTool scripts. See the UserDocumentation for more details.
Python has several benefits as a platform for our package. It is an open-source language, enthusiastically developed by a large community of people drawn from both academic and commercial backgrounds. The powerful object-oriented and dynamic typing features of the language greatly aid in the development of flexible and intuitive data structures and user interface (UI) elements. Being a high level language its interactive mode of UI, via a prompt, follows the same principle as that of Matlab, and shares the same advantages for quick prototyping, querying and scripting of complex object manipulations and computations.
For more about what Python has to offer in scientific computing see the PythonResources page or a page at the python wiki.
In our design we have emphasized modularized data structures and interface design that facilitates data-driven approaches to the modeling of physical processes, and we have built upon standard numerical, scientific and graphics libraries for Python (for instance, SciPy and MatPlotLib). These, in turn, make use of well-established and efficient legacy codes for numerical integration of ODEs, and for dealing with linear algebra, optimization, and root solving (for instance, the LAPACK and MINPACK Fortran libraries). These legacy codes are typically interfaced using SWIG. The low-level languages of these codes provide the computational speed that Python itself lacks, in the places for which computation is most intensive.
All of the code involved in the PyDSTool project is open source, and we have aimed to create as few dependencies on external software packages as possible. In particular, the package can be used with Microsoft Windows, Mac OS X and Linux machines.
On top of the third-party libraries we have added several new tools and capabilities. We have enhanced legacy numerical integration code for ordinary differential equations to perform various additional tasks of use in hybrid systems modeling, implemented at the C-code level for maximal efficiency. This includes supporting discrete event detection during dynamical evolution. Adding arbitrary user-specified event detection to a model permits ODEs and maps to be used in combination as "hybrid" dynamical systems. Also, the inclusion of data-based time series inputs to a dynamical system’s evolution equations is a feature that aids data-driven modeling.
Utilities have been added that allow the movement of data and model specifications both in and out of PyDSTool, for sharing in other software environments. As well as basic importing and exporting of numerical data as text files, this also includes more systems-level interfacing. For instance, a user can export a dynamical model’s definition to a Matlab environment in which Automatic Differentiation is available for parameter sensitivity calculations (via the package ADMC++). Also, PyDSTool can be interfaced with the systems biology modeling package SloppyCell, through which PyDSTool inherits access to an interface with the Systems Biology Markup Language (SBML) for model specification, and the LaTeX mathematical markup language. Further interfaces to packages are in active development, such as to the original DsTool and to other simulation tools such as XPP or NEURON.
Support for the CUDA Graphics Processing Unit parallel computing system is nearly complete (March 2012). Also, there will be better support for interfacing with the SBML model markup language (soon, via PySCes) and the NineML neural system markup language (less soon). More details when they come!
Support for delayed systems is currently limited to dirac delta function pulses, as part of a hybrid system (see HybridSystems). DDE integrators written by Hairer and Wanner follow the same interfacing pattern as our existing integrators and will be added later.
Automatic differentiation and Taylor-series integration capabilities will be added once we can get a working interface to an open-source AD package such as ADOL-C. In the meantime, we provide a model export tool to interface PyDSTool models to work with the ADMC++ environment in Matlab, a package that was also developed at Cornell (by Eric Phipps). See this paper about ADMC++ and its application to parameter estimation for periodic orbits.
Although adequately supported in the underlying numerics of Python and SciPy, PyDSTool does not presently support dynamical systems having phase spaces ranging over the complex numbers. The application of PyDSTool to date has been to certain types of model in the physical sciences which have not required expressions of complex dynamics. It is hoped that at some point this can be redressed with explicitly complex dynamical systems. In the meantime, complex dynamics must be represented using two-dimensional real number phase spaces, and complex arithmetic implemented using auxiliary user-defined functions on 2D reals.
We have successfully tested an update of our automatic generation and compilation of C-code, although it does not yet avoid Python's distutils package by using Scons or autoconf / automake. This updated improves the efficiency of C-based vector field support and will make them more easily configured in new installations of Python/NumPy/SciPy/Matplotlib/PyDSTool.
We look forward to adding a range of other standard tools that are available in other packages for dynamical systems, Poincare maps, support for symbolic dynamics and complex dynamics, BVP solvers, averaging tools, invariant manifold computations, and so on.
The DsTool package was written at Cornell University in the early 1990s under the direction of Prof. John Guckenheimer, and has provided many years of functionality to the applied mathematics community. In about 2005, a group of us at Cornell led by Prof. Guckenheimer decided to write a new package from the ground up, as our concept of what is needed in our community has evolved. Our new project is different from existing approaches because it attempts to combine a large array of tools under one, centralized umbrella. We wish to support both straightforward simulation tools, such as numerical integrators of differential equations, and sophisticated analysis tools such as continuators and parameter estimation / sensitivity tools.
Individually, several examples of the software tools to implement these algorithms exist in the world of scientific computing. However, there has not yet been a one-stop solution to providing mathematically sophisticated users with a combined suite of such tools that are highly integrated and interactively operated. For instance, Matlab is geared much less towards applied mathematicians in this regard as it is to physicists and engineers. It is still typical for applied scientists to need to use a variety of separate programs to achieve their dynamical systems modeling goals, but increasingly sophisticated questions are being asked of the models, which are ever-harder to answer when the tools are not integrated. Example domains that will benefit from PyDSTool are computational neuroscience, biomechanics and robotics, and advanced engineering projects.
In broad terms, PyDSTool combines or extends a range of features from existing packages into an integrated and open environment for modeling complex dynamical systems in scientific applications. There are several existing software packages for working with dynamical systems. There are a variety of features to compare packages by. Some of the major ones are:
PyDSTool fills a clear niche in the space of features compared to all existing packages known at the time of writing. This is summarized in the following chart, which is compiled to the best of the author's knowledge and in places may be somewhat subjective (please send any corrections to me by email). An "X" means the feature is supported. The abbreviation "CLI" means Command Line Interface (interactive), "GUI" means Graphical User Interface, and "DIY" means Do It Yourself! Some of the packages in the table are discussed below.
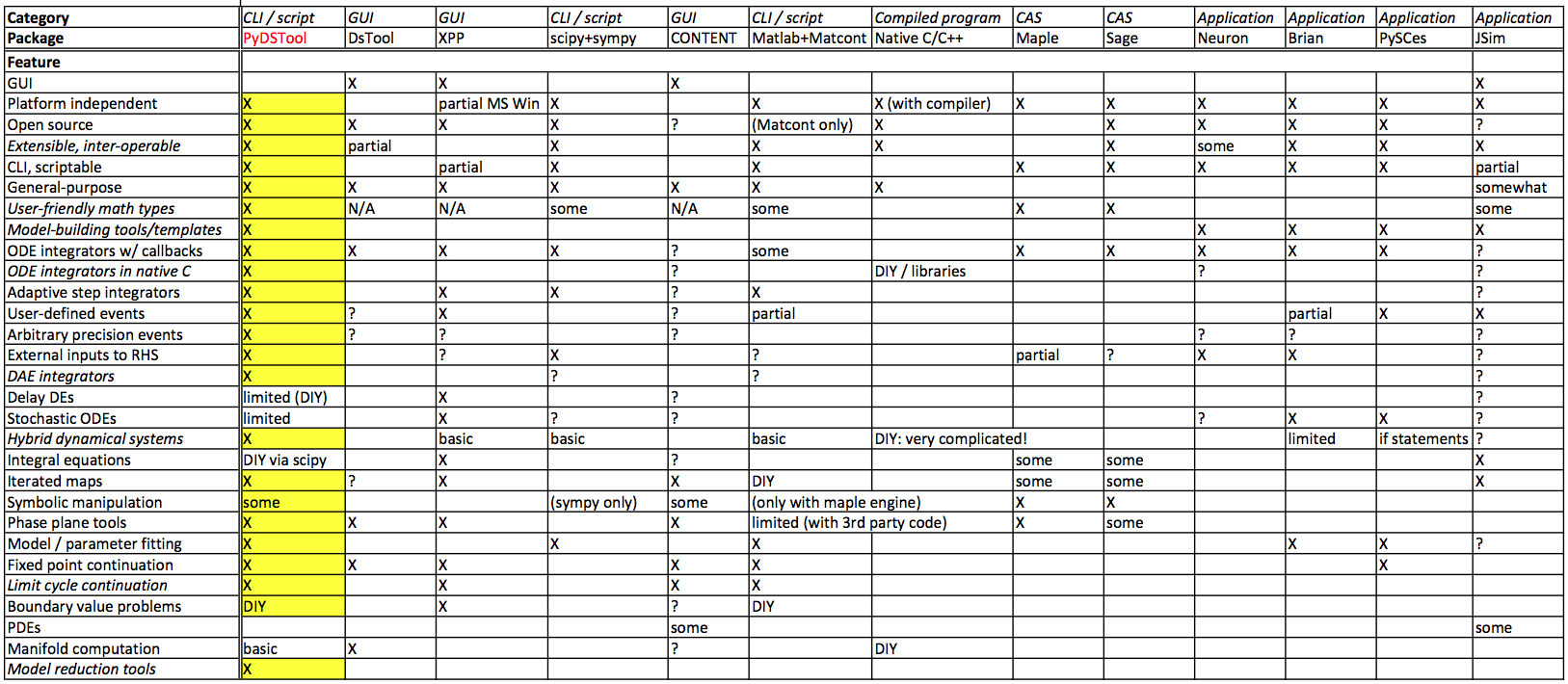
PyDSTool provides dynamical systems-specific tools that Matlab does not.
Other science and data analysis tools are present in the working environment of PyDSTool through access to Scipy, Numeric, and Pylab. Many of these are ultimately imported from the same standard C or Fortran libraries that Matlab uses.
PyDSTool retains many of the advantages of the interactive environment that is Matlab's shell.
PyDSTool's numerical integrators are much faster and more richly-featured than Matlab's and support a wider range of ODE, DAE, DDE and discrete map models.
PyDSTool model building scripting language and "index free" data structures make simulations and model exploration easy.
PyDSTool provides no graphical interface to its data structures.
See also this comparison of the NumPy library to Matlab.
XPP (X-PhasePlane)
PyDSTool supports arbitrarily large systems, and supports a representation of their inherent structure through our compositional model-building scripting language. This facilitates building such large models.
PyDSTool supports a wider range of differential equation right-hand side expressions, except for stochastic systems.
PyDSTool supports long names (more than 9 characters).
PyDSTool's continuation and bifurcation analysis is more closely integrated with core PyDSTool tools and data structures, and is scriptable.
PyDSTool's simulations and analysis can be embedded inside other algorithms, and automated using interactive or "intelligent" scripts. This is because PyDSTool is an open and programmatically-based environment.
XPP supports general delay-differential systems, a wider variety of ODE integrators, and a BVP solver.
XPP provides a fully-graphical user interface, postscript output, visualization tools for arrays, a GUI for a subset of AUTO, and a lot of other good stuff.
PyDSTool offers a suite of common continuation tools (via PyCont), but cannot yet be considered comprehensive.
PyDSTool/PyCont is an "open environment", like Matcont. This facilitates data exchange between different packages.
PyDSTool offers closer integration of continuation with the rest of the working environment.
PyCont is written in pure Python (except for the interface to AUTO), and is unlikely to work as fast as AUTO, although it is competitive with Matcont. The dynamically-linked C code for periodic orbit continuation re-uses components of AUTO directly. Because we create a C-code version of a vector field, PyCont+AUTO is technically a little faster than the most recent full native release of AUTO for continuation of periodic orbits, which uses Python callbacks to evaluate user-supplied functions.
PyDSTool provides a centralized, general purpose environment.
Python has a strong object-oriented capability.
PyDSTool utilizes automatically-generated and dynamically-linked C code for fast execution of user-defined vector fields. Hand-written modifications to this C code, or externally written DLL code can also be utilized.
Python's dynamic typing makes embedding PyDSTool objects inside other Python computations very simple to set up.
Python's interactive environment (i.e. interpreted language) is excellent for exploration of a mathematical system, and introspection of the associated PyDSTool objects.
SUNDIALS, PySUNDIALS, odeint. These are low level codes for solving ODEs, DAEs, etc.
Sage is an open-source and free mathematics system for Python, styled somewhat like Maple. Along with Mathematica, and others, these are Computer Algebra Systems (CAS) primarily.
Neuron at Duke, Neuron at Yale, Genesis, and Brian. These are biophysical neural network simulators.
A PyDSTool toolkit for compartmental modeling provide templates of data structures and utilities for sophisticated hierarchical and composable model building, using a modern object-oriented model specification language built in to PyDSTool (including symbolic expression handling).
The PyDSTool templates are not comprehensive, and are not as fully-featured as some of those designed for use with Neuron or Genesis. We are working to broaden the support for different types of model. A tutorial is available for PyDSTool's computational neuroscience templates.
PySCes simulator for cellular systems is an open source package.
PyDSTool source code is hosted by SourceForge: